Universal Antibiotic Resistance Genes

55 Genetic Elements Associated With Resistance to 10 Different Classes of Antibiotics
A new and improved antibiotic resistance genes panel has been released by Diagnostic Solutions Laboratory. Useful for patients who have been hospitalized, treated with antibiotics, or who have stubborn, chronic infections, the Universal Antibiotic Resistance Genes panel will tell clinicians if antibiotic resistance is a potential stumbling block for their patient. The Universal Antibiotic Resistance Genes panel can be ordered as an add-on to the GI-MAP or to the GI-Pathogens panel. It detects the presence of 55 genetic elements associated with resistance to 10 different classes of antibiotics.
Multidrug resistance is a significant public health threat worldwide.1 The Centers for Disease Control estimate 35,000 deaths in the United States due to antibiotic-resistant infections each year and the treatment of antibiotic resistant microbes totals more than $4.6 billion in annual health costs. In response, antimicrobial stewardship programs are being promoted to combat drug resistance, treat infections more efficiently, protect patients from harm of unnecessary antibiotics, and reduce healthcare costs.4 Integrative and functional medicine clinicians can support antibiotic stewardship by incorporating the findings from the Universal Antibiotic Resistance Genes into their patient protocols.5
Both Gram-negative and Gram-positive bacteria can evade antibiotic therapy by carrying specialized mobile genetic elements on plasmids (circular DNA molecules in the cytoplasm). These mobile elements can be transmitted through horizontal genetic exchange between bacterial cells, and even between different species. Horizontal genetic exchange can be thought of as bacteria swapping useful genes to promote their survival. This genetic exchange of mobile elements plays a central role in the acquisition and spread of antibiotic resistance in bacterial populations.1
The Universal Antibiotic Resistance Genes panel detects genes associated with resistance to the most commonly prescribed antibiotics for gastrointestinal infections. The mobile genetic elements in the panel can be found in a variety of different microbes. The presence of these genes in a bacterial population have been associated with moderate to high levels of antibiotic resistance in human gastrointestinal infections.
Universal Antibiotic Resistance Genes Panel – Identifies Microbial Resistance to These Classes of Antibiotics
- β-lactams
- Fluoroquinolones
- Vancomycin
- Macrolides
- Ciprofloxacin
- 5-Nitroimidazoles (non-Helicobacter pylori)
- Trimethoprim
- Sulfonamides
- Methicillin
- Chloramphenicol
Understanding the Report
For most antibiotic classes, there are several different genes that bacteria can use to evade the antibiotic. The gene type is dependent on the mode of resistance and the organism(s) in which it may be found. Number and letter combinations are given to genes involved in antibiotic resistance. For example, vanA1, vanA2, and vanB are genes that provide resistance to vancomycin, usually found in Enterococcus species.2 Use the acronym and the antibiotic class to further research each gene in the medical literature.

Mechanisms of Antimicrobial Resistance
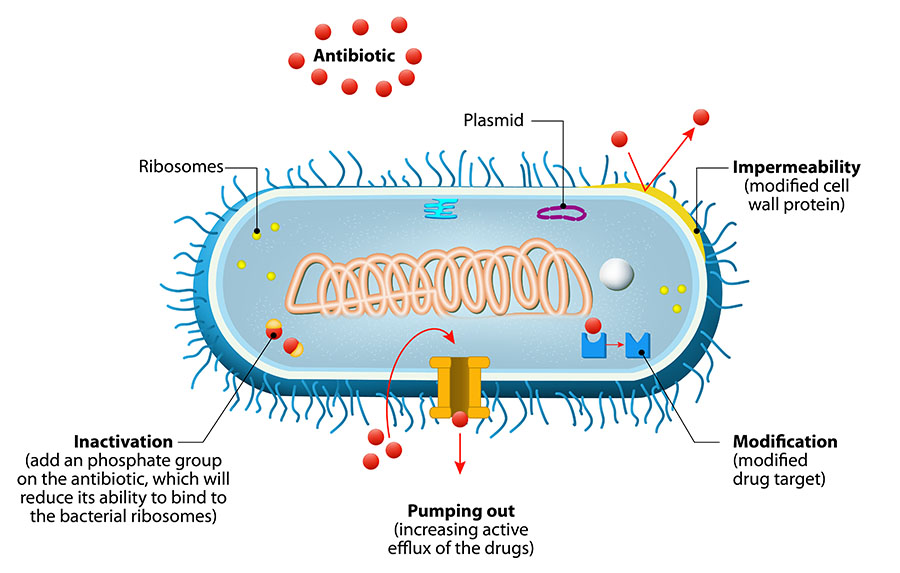
Figure 1. Bacteria avoid antibiotic activity using these basic mechanisms: (1) decrease uptake of the drug (impermeability), (2) modify the drug inside the cell so that it cannot bind, (3) pump out the drug with an efflux pump, and/or (4) modify the target of the drug so that the antibiotic cannot bind.3
"Universal" Resistance Genes are Clinically Useful
Antibiotic resistance genes are "universal" because these genetic elements are not specific to a single bacterial species. Therefore, findings may pertain to all microorganisms found in the patient's fecal sample. In other words, the Universal Antibiotic Resistance Genes panel may show a positive for vancomycin resistance, but it does not determine if Enterococcus faecalis on the report is specifically resistant to the vancomycin antibiotic.
Even though the Universal Antibiotic Resistance Genes results are not specific to a single bacterial species, they are clinically actionable. When an antibiotic resistance gene is detected, it indicates that population(s) of bacteria have resistance to that class of antibiotics. It is, therefore, not advisable to utilize this class of antibiotics as the first choice in an antibiotic protocol. In addition, as different species of bacteria can rapidly share DNA via horizontal gene transfer, the presence of antibiotic resistance in any bacterial population is reason enough to avoid use of that drug class. A resistance gene is a threat to the entire microbiome.
Antibiotic therapy can be better tailored to each unique patient when the antibiotic resistance profile of his or her microbiome is known. Multiple antibiotic resistance genes could explain gastrointestinal infections that do not respond to – or are made worse by – antibiotic treatment. Herbal antimicrobials, which are commonly used in integrative and functional medicine, may be ideally suited to patients with multiple antibiotic resistance genes. Plant extracts are less susceptible to antibiotic resistance.6

Author Cass Nelson-Dooley, MS
Cass Nelson-Dooley, MS, studied medicinal plants in the rain forests of Panama, in 2003 as a Fulbright Scholar, and then launched a career in science and natural medicine. She researched the pharmacology of medicinal plants at the University of Georgia and AptoTec, Inc. She has over 15 years of experience teaching doctors about integrative and functional laboratory results.
At Diagnostic Solutions Laboratory, she analyzes GI-MAP stool tests and develops educational tools. Ms. Nelson-Dooley owns Health First Consulting, LLC, a medical writing, patient education, and consulting firm that serves the integrative and functional medicine industry. Ms. Nelson-Dooley is the author of Heal Your Oral Microbiome and has published case studies, book chapters, and journal articles about natural medicine, nutrition, and laboratory testing.
REFERENCES
- Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clinical microbiology reviews. Oct 2018;31(4)doi:10.1128/CMR.00088-17
- Ahmed MO, Baptiste KE. Vancomycin-Resistant Enterococci: A Review of Antimicrobial Resistance Mechanisms and Perspectives of Human and Animal Health. Microb Drug Resist. Jun 2018;24(5):590-606. doi:10.1089/mdr.2017.0147
- Antibiotics in Our Water Supply - Are We Polluting the Element of Life? ThermoFisher Scientific. Accessed August 8, 2023. https://www.thermofisher.com/blog/behindthebench/antibiotics-in-our-water-supply-are-we-polluting-the-element-of-life/
- Bankar NJ, Ugemuge S, Ambad RS, Hawale DV, Timilsina DR. Implementation of Antimicrobial Stewardship in the Healthcare Setting. Cureus. Jul 2022;14(7):e26664. doi:10.7759/cureus.26664
- Watkins RR. Antibiotic stewardship in the era of precision medicine. JAC Antimicrob Resist. Jun 2022;4(3):dlac066. doi:10.1093/jacamr/dlac066
- Álvarez-Martínez FJ, Barrajón-Catalán E, Micol V. Tackling Antibiotic Resistance with Compounds of Natural Origin: A Comprehensive Review. Biomedicines. Oct 11 2020;8(10)doi:10.3390/biomedicines8100405
- Ramadan AA, Abdelaziz NA, Amin MA, Aziz RK. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase-producing Escherichia coli. Scientific reports. Mar 12 2019;9(1):4224. doi:10.1038/s41598-019-39730-0